For the primary time, researchers have efficiently extracted and decoded RNA from an extinct animal.
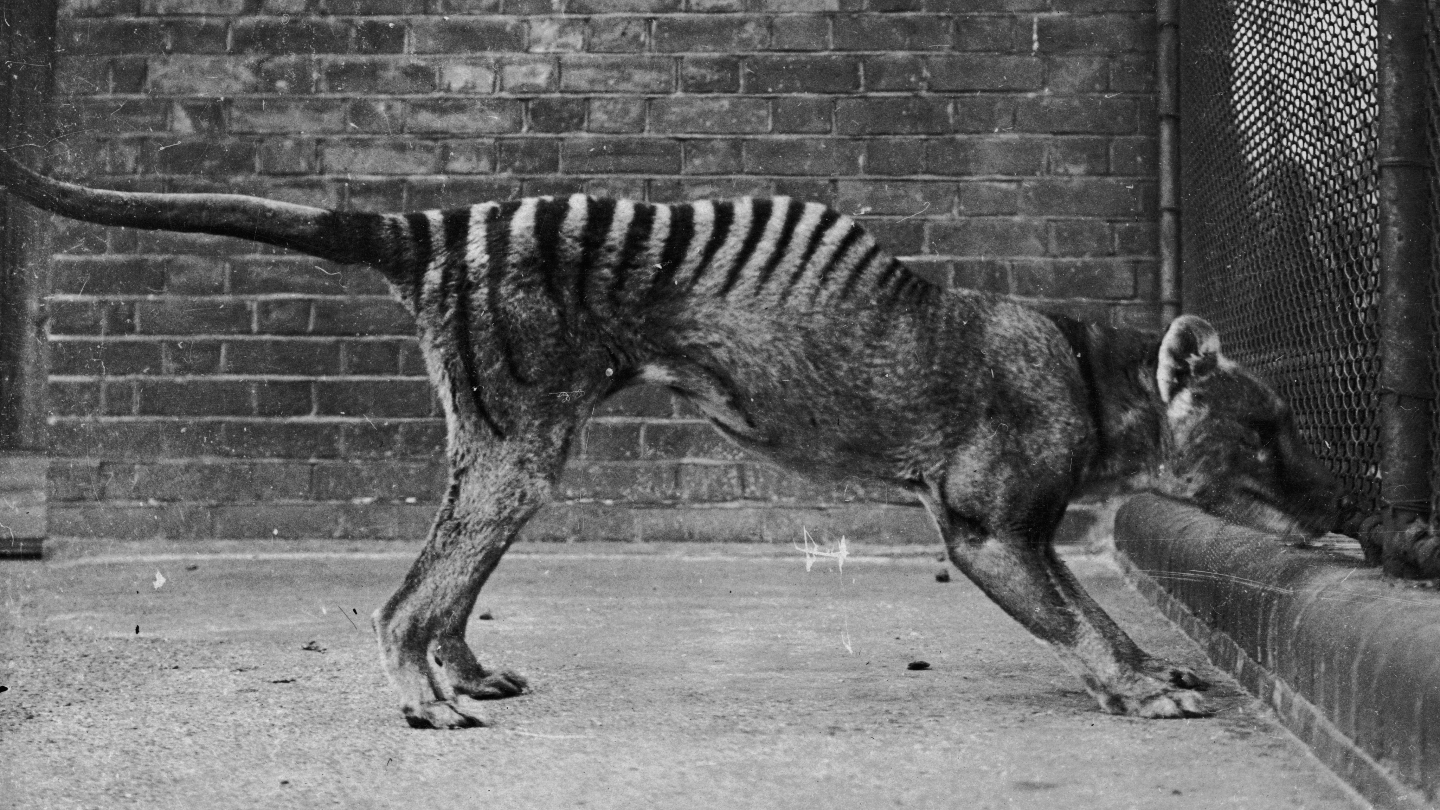
The thylacine, often known as the Tasmanian tiger, was a wolflike marsupial that went extinct after the final one died in a zoo in Hobart, Tasmania in 1936. Now a roughly 130-year-old museum specimen has yielded bits of RNA, the delicate molecules chargeable for turning DNA’s genetic directions into mobile capabilities, researchers report within the August Genome Analysis. The outcomes shed new gentle on thylacine biology and should inform efforts to deliver the marsupial again from extinction.
With darkish stripes operating over its tawny coat from its shoulders to its tail and jaws able to opening greater than 80 levels, the thylacine (Thylacinus cynocephalus) was a hanging animal. However the carnivores have been no match for people: As sheep farming proliferated within the 1800s in Tasmania — the house of the final remaining wild inhabitants of thylacine — the animals have been continuously implicated in killing livestock. Within the late nineteenth century, a bounty was established for each grownup thylacine killed, and the animals have been hunted practically to extinction.
Lately, researchers have mapped out the thylacine genetic blueprint, along with the genomes of different extinct animals just like the woolly mammoth (SN: 2/17/21). However such investigations have been all centered on DNA. Solely RNA can reveal how an organism’s cells really functioned, says Emilio Mármol-Sánchez, a geneticist on the Karolinska Institute in Stockholm. “You see the actual biology of the cell.”
In 2020, Mármol-Sánchez and colleagues got here throughout a thylacine specimen in storage on the Pure Historical past Museum in Stockholm. “It was simply there in a cabinet,” says Mármol-Sánchez, then at Stockholm College and the Heart for Paleogenetics in Stockholm.
The crew collected six small samples of pores and skin and muscle from the desiccated animal. Again within the laboratory, the researchers floor every pattern right into a powder and added chemical substances that remoted nucleotides, the constructing blocks of RNA. Subsequent, the crew used a pc algorithm to check these strings of nucleotides, or sequences, with a database containing the genomes of 1000’s of animals, crops, fungi, micro organism and viruses — together with the thylacine.
The crew concluded that roughly 70 % of the RNA sequences they discovered have been reliably thylacine, with some contamination from human RNA for the reason that thylacine specimen was repeatedly dealt with.
Their evaluation revealed completely different protein-coding RNA molecules of their pores and skin and muscle samples. That is sensible, Mármol-Sánchez says. “Muscle cells and pores and skin cells serve fairly completely different capabilities within the physique.” For example, the researchers pinpointed RNA molecules that coded cells to make slow-twitch muscle fiber, which helps with endurance.
The crew additionally discovered over 250 thylacine-specific brief RNA molecules often called microRNAs. These RNA sequences regulate cell functioning, Mármol-Sánchez says. “They’re the policemen of the cell.”
These are spectacular outcomes, says Andrew Pask, a developmental biologist on the College of Melbourne in Australia who was not concerned within the analysis. Many researchers by no means even search for RNA, he says. “It’s a lot much less steady than DNA.” And the findings are doubly spectacular on condition that the specimen was saved at room temperature, Pask says, reasonably than in sterile or frozen circumstances. (RNA has been beforehand extracted from samples of present species preserved in alcohol or ice.) “It’s reworked the best way that we take a look at museum and archive specimens.”
Within the not-too-distant-future, Pask and different researchers are hoping to deliver thylacine again to Tasmania. Their plan to de-extinct the animal includes modifying the genes of one of many thylacine’s closest dwelling family, one other marsupial referred to as a fat-tailed dunnart (Sminthopsis crassicaudata). These new findings may very nicely inform that effort, Pask says, by revealing genes that managed the animal’s attributes. “It’s an entire different layer of data.”